RNALOC
Granted by ANR
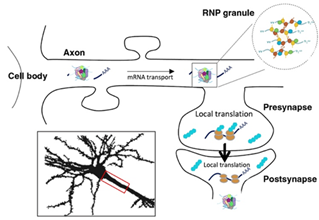
Subcellular targeting of RNA molecules has emerged as a prominent mechanism enabling tight spatiotemporal control of protein production. RNA localization is particularly important in neuronal cells, which extend long axonal and dendritic branches that must locally respond to the activity of their partners. While classical transcriptomic studies have been performed on isolated cell fractions, we still have a very limited understanding of i- the relevant subcellular scales of RNA compartmentalization (axon-wide vs synapse specific) and ii-the physiological cues controlling this process in vivo. Using the power of Drosophila, and exciting preliminary results uncovering the existence of physiologically-relevant, cue-induced, new levels of RNA compartmentalization in neuronal cells, we propose to dissect the mechanisms underlying RNA compartmentalization, and to develop cutting edge approaches to generate spatially-resolved 3D atlases of RNA distribution in distinct neuronal populations and different conditions in vivo. Combining the complementary expertise of the 3 partners involved (F. Besse, expert in spatial RNA biology in Drosophila; X. Descombes, specialist in advanced image analysis and K. Lebrigand, bio-informatician expert in spatial transcriptomics), we will specifically study the cellular machineries involved in the local recruitment and anchoring of selected RNAs, and unravel the importance of activity cues (Aim 1). We will also develop an innovative spatial transcriptomics pipeline to cartography in vivo, with unprecedented resolution, and at a transcriptome-wide scale, RNA subcellular distribution in complementary cell types and compartments (Aim 2). This approach will be further used to assess the global impact of transport and anchoring machineries and the contribution of neuronal activation.
With this highly interdisciplinary project, we will thus explore new dimensions (3D, transcriptome-scale) and new processes (compartment-specific targeting, control by external cues) underlying subcellular trafficking of RNA molecules in highly polarized neuronal cell types in vivo.
With this highly interdisciplinary project, we will thus explore new dimensions (3D, transcriptome-scale) and new processes (compartment-specific targeting, control by external cues) underlying subcellular trafficking of RNA molecules in highly polarized neuronal cell types in vivo.
