Spatial imaging-based transcriptomics
Author: Kévin Lebrigand
Evolution of spatial transcriptomics
The field has quickly expanded in recent years, and several new technologies have been developed that all aim to combine gene expression data with spatial information. The vast array of methodologies displays fundamental differences in their approach to obtain this information, and thus, demonstrate method-specific advantages and shortcomings.
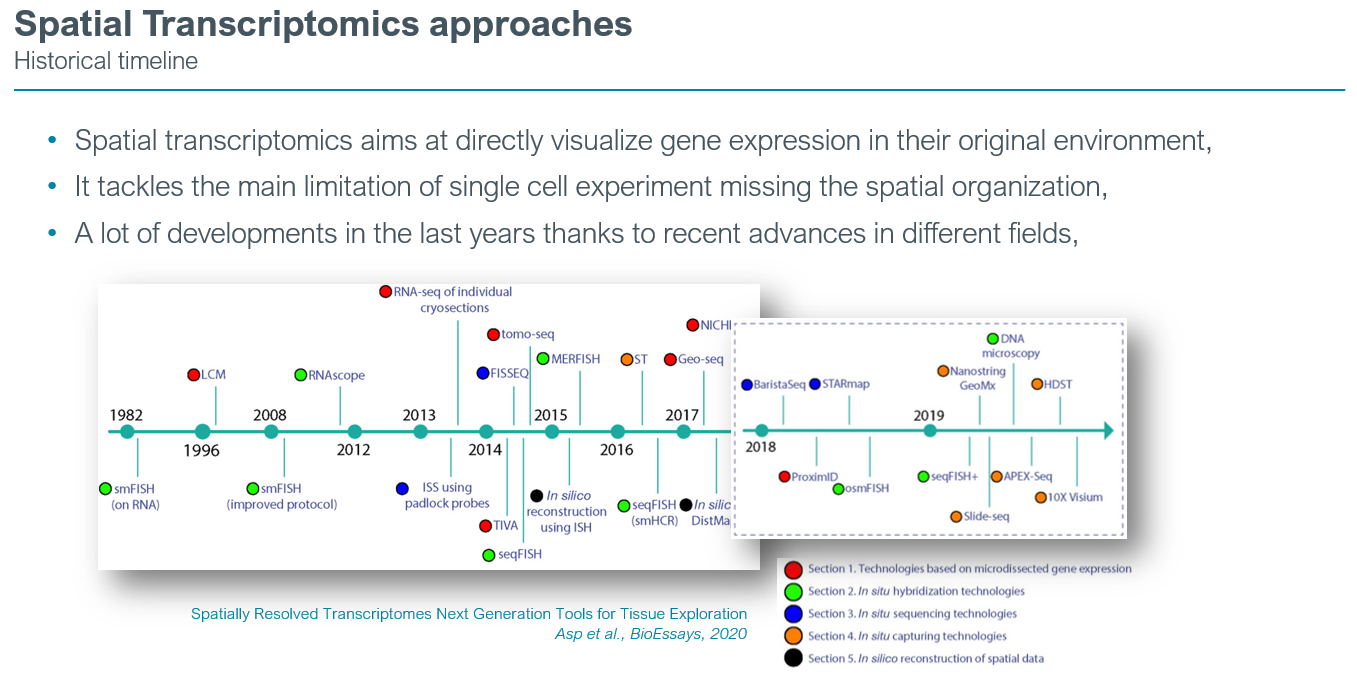
In the last 5 years most of the spatial transcriptomics paper have utilized in-situ capture methods such as the Visium assay (10x Genomics). Based on oligo-dT barcodes primers of 55µm diameter, the signal is basically obtained from 5 to 15 cells under each spots. The polyA capture renders the Visium assay whole transcriptome. Since 2 years the field is rapidly evolving towards the recovery of the single-cell resolution and even better the sub-cellular resolution thanks to imaging-based acquisition. Those assays are limited to a restricted number of gene targets that could be image, typically between 300 and 1000.
Spatially Resolved Transcriptomes Next Generation Tools for Tissue Exploration
Technologies
Single-molecule spatial imaging-based transcriptomics via multiplexed RNA fluorescent hybridization can reveal detailed tissue organization at the single-cell resolution. Currently 3 platforms are accessible to the academics community : CosmX (nanostring), Merscope (Vizgen) and Xenium (10x Genomics). Those 3 platforms utilizes different efficient chemistries to revealed the sub-cellular localization of transcripts molecules.

At IPMC we choose in october 2022 to acquire the Merscope system (Vizgen) and plan an upgrade in march 2024 with the acquisition of the Xenium (10x Genomics) that shows higher performances compared to the merscope in FFPE tissue slices. We envision to perform fresh frozen sample using the Merscope and FFPE sample with the Xenium system in 2024.
Systematic comparison of sequencing-based spatial transcriptomic methods
A Comparative Analysis of Imaging-Based Spatial Transcriptomics Platforms
Planning a Merscope/Xenium spatial transcriptomics experiment
Defining the gene target panel
One of the critical task that require a careful attention is the selection of the target gene panel that will be imaged during the chemistry. This target gene panel needs to integrate genes that will have different mission during statistical analysis that will be performed by bioinformaticians after acquisition on the system: (i) first you will need genes for the cell typing process (assign a cell type) for all cells that might be on your slices, thanks to global initiative such as the Human Cell Atlas, a lot of single-cell resources exist today to compose this list; (ii) second, you will integrate genes that are part of pathways that you want to explore in your system; (iii) you might want to integrate genes that acts as ligand-receptor pairs in your systems. Note that compagnies provide several pre-designed panels related to the global aim of your study objective (Cancer panel, brain panel, )
Access vizgen custom panel creator
Vizgen predesigned cancer and neuronal gene panels
Access Xenium predesigned panels
Note that the Xenium platform allows to complement pre-designed panels with custom gene panels (50 or 100 custom gene). Note that custom panel can be delivered by both companies within 5-6 weeks after placing the order.
Tissue slices and experimental design
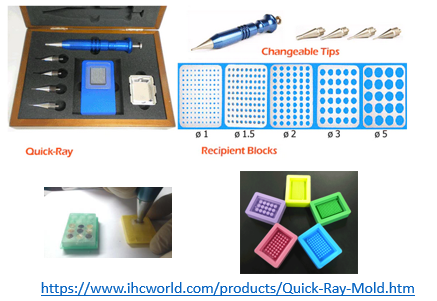
Imaging area are large (ie. 10x10mm for vizgen, 10x22mm for Xenium) which allows to image several pieces of tissue per flow cells (ie. 1 for vizgen, 2 for Xenium). Take into consideration to use Tissue Microarray Assays (TMA) to increase the number of samples and replicates and decrease cost of experiment. Sample cores can be punch and place manually using a “Quick-Ray Manual Tissue Microarrayer Full Set” and “Quick-Ray Molds”:
Both systems allow for FFPE and Fresh frozen samples to be processed. You will need to have access and knowledge about sectioning (5-10 µm) block using a microtome.
Vannan et al., Biorxiv, 2023, method section from Banovich’lab on lung fibrosis
"As Xenium in Situ technology examines RNA, all protocol workstations and equipment were cleaned using RNase Away (RPI 147002) followed by 70% Isopropanol. All reagents, including water, were molecular grade nuclease-free. Sample preparation began with rehydrating and sectioning FFPE blocks on a microtome (Leica RM2135). 5um sections were placed onto Xenium slides (10X Genomics). Following overnight drying, slides with placed samples were stored in a sealed desiccator at room temperature for ≤10 days. Slides were then placed in imaging cassettes for the remainder of the preparation. Tissue deparaffinization and decrosslinking steps made sub-cellular RNA targets accessible. Gene panel probe hybridization occurred overnight for 18 hours at 50°C (Bio-Rad DNA Engine Tetrad 2). Subsequent washes the next day removed unbound probes. Ligase was added to circularize the paired ends of bound probes (2 hours at 37°C) and followed by enzymatic rolling circle amplification (2 hours at 30°C). Slides were washed in TE buffer before background fluorescence was chemically quenched; autofluorescence is a known issue in lung tissue as well as a byproduct of formalin-fixation 50. Following PBS and PBS-T washes, DAPI was used to stain sample nuclei. Finalized slides were stored in PBS-T in the dark at 4°C for ≤5 days until being loaded onto the Xenium Analyzer instrument. Stepwise Xenium FFPE preparation guidelines and buffer recipes can be found in 10X Genomics’ Demonstrated Protocols CG000578 and CG000580."Spatial transcriptomics data analysis
Spatial transcriptomics assays required important bioinformatics resources to permit efficient exploration nad final production of figure for publication. Please consider securing bioinformatics resources at the begining of your project. Several statistical data analysis workflows exists to handle Xenium and Vizgen data :
Seurat v5, Satija lab (New York Genome Center, NY, USA
Squidpy-Scanpy-Anndata, Theis lab (Helmholtz Center Munich, Germany
Monkeybread, Immunitas Therapeutics
Giotto, Dana-Farber Cancer Institute , Boston, USA
SpatialData, Oliver Stegle, European Molecular Biology Laboratory (EMBL), Heidelberg, Germany
